Modules and scripts#
# For working with paths
from pathlib import Path
# Usual imports
import numpy as np
import matplotlib.pyplot as plt
import nibabel as nib
We get a 4D volume to work on:
# Load the function to fetch the data file we need.
import nipraxis
# Fetch the data file.
data_fname = nipraxis.fetch_file('ds107_sub012_t1r2.nii')
# Show the file name of the fetched data.
data_fname
'/home/runner/.cache/nipraxis/0.5/ds107_sub012_t1r2.nii'
Let’s say we have a function that loads a 4D image, and returns a list with mean values across all voxels in each volume:
def vol_means(image_fname):
img = nib.load(image_fname)
data = img.get_fdata()
means = []
for i in range(data.shape[-1]):
vol = data[..., i]
means.append(np.mean(vol))
return np.array(means)
For extra points: can you work out a better way to do the same thing without a loop?
First we check the vol_means function works:
my_means = vol_means(data_fname)
plt.plot(my_means)
[<matplotlib.lines.Line2D at 0x7fa19c04c370>]
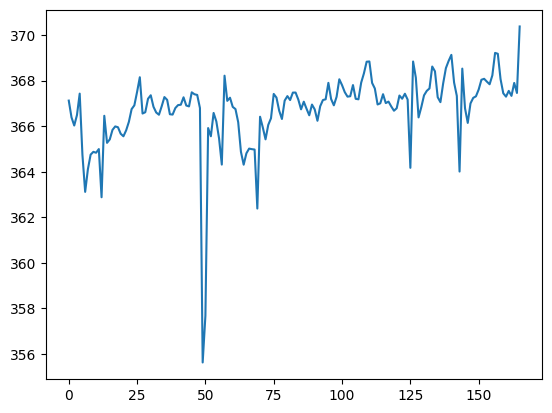
We are interested in outlier detection. Let’s make a routine to detect outliers:
def detect_outliers(some_values, n_stds=2):
overall_mean = np.mean(some_values)
overall_std = np.std(some_values)
thresh = overall_std * n_stds
is_outlier = (some_values - overall_mean) < -thresh
return np.where(is_outlier)[0]
outlier_inds = detect_outliers(my_means)
outlier_inds
array([ 6, 12, 49, 50, 69])
my_means[outlier_inds]
array([363.11401367, 362.87746233, 355.6257952 , 357.68177316,
362.3819894 ])
These seem like useful functions.
We decide we’d like to use these in another notebook.
Off we go and copy / paste these functions into another notebook
Go there, and have a look at how it does on some more data.
Introducing modules#
Over in another notebook, we found that the there is a mistake in the copy/pasted code.
Of course we could fix it here, and then go and fix it in the other notebook.
But, a better way, is:
Put the code in one place, for both notebooks to use
Fix it in one place, and
Test the code! (More on this later).
A Python module is a Python file with functions (and variables and other
things) that other modules and scripts may use. These other modules and scripts
will import the module.
The simplest possible module is just a file with a .py extension, with
Python code.
In the cell below, we are using the notebook machinery to write some context
to a text file called volmeans.py in the current working directory, with a
%%file Jupyter magic command at the top of the cell.
First we check whether we already have a volmeans.py in the current
directory, using pathlib:
Path('volmeans.py').is_file()
True
That should show False meaning that the file does not exist.
Usually we would go on to write the file with our text editor. You will very
rarely need or use the %%file magic; it is really only useful for
demonstration notebooks like this one.
%%file volmeans.py
""" File to calculate volume means, detect outliers
"""
import numpy as np
import nibabel as nib
def vol_means(image_fname):
""" Calculate volume means from 4D image `image_fname`
"""
img = nib.load(image_fname)
data = img.get_fdata()
means = []
for i in range(data.shape[-1]):
vol = data[..., i]
means.append(np.mean(vol))
return np.array(means)
def detect_outliers_fixed(some_values, n_stds=2):
overall_mean = np.mean(some_values)
overall_std = np.std(some_values)
thresh = overall_std * n_stds
is_outlier = np.abs(some_values - overall_mean) > thresh
return np.where(is_outlier)[0]
Overwriting volmeans.py
We now do have a file in the current directory called volmeans.py:
Path('volmeans.py').is_file()
True
We can use this new module very simply - by importing it, as we have
imported other modules and libraries:
import volmeans
As with other modules, you can explore the functions and data available in this module by making a new cell, and typing volmean. followed by the tab key.
# Uncomment to try exploring the volmeans module.
# volmeans.
Now we find that the functions defined in the module are attached to the imported module, volmeans.
means_again = volmeans.vol_means(data_fname)
volmeans.detect_outliers_fixed(means_again)
array([ 6, 12, 49, 50, 69, 165])
Changing the module, reloading#
Usually, when we write a module, we import it into the notebook and use it, without need to change the module.
But sometimes, if you are developing interactively, you find you want to edit the module file and rerun the code in the module.
Let us simulate that process now, by writing a new, very simple module file,
with a print at the end of the module.
%%file simplemod.py
def double_me(a):
return a * 2
print('Result of double_me on 2', double_me(2))
Writing simplemod.py
Now, when we import the module, Python executes the code in the module, which first, defines the double_me function and then prints the result. So, when we import, we see the printed result, like this:
import simplemod
Result of double_me on 2 4
Now let’s imagine we are still in the same Python session. For example, imagine, as is the case for us now, we are using the same Jupyter kernel. But we want to rewrite the module to print out the result for a more interesting number.
Note We would normally edit the module in our text editor, but here we’re
using %%file again to simulate that process of editing and writing the file:
%%file simplemod.py
def double_me(a):
return a * 2
print('Result of double_me on 13', double_me(13))
Overwriting simplemod.py
Then we run the import again.
# Second time we import the module.
import simplemod
Oh dear - Python hasn’t picked up the new version of the file. In fact, it isn’t printing anything - it isn’t running the print statement at all — from the previous version or from the new version.
This is due to the mechanism Python uses to import the file. When Python
does an import, it first tries a shortcut, and only if that fails, does it
load and run the code from the file. The process is:
Shortcut: Python first checks if it already has the defined module, and returns that if so, otherwise;
Long way round: Python reads the text file, runs the code inside, and returns the resulting module.
In the case above, our second-time-round import simplemod triggers the
shortcut, where Python already has the module, so it returns the code from
that module, without running it again. That is the code that existed the
first time we did import simplemod, so we get the result from the initial
long way round load of the original code.
Usually Python’s process is what we want, because we usually don’t find ourselves editing Python modules after we have imported them, in the same Python session. When we haven’t edited the module, the shortcut saves a lot of time, when we import the same module several times.
For the rare case where we do edit the module while the Python process is running we can force Python to go the long way round, like this:
# Library to execute the importing logic.
import importlib
# Trigger long-way-round import of module.
importlib.reload(simplemod)
Result of double_me on 13 26
<module 'simplemod' from '/home/runner/work/textbook/textbook/simplemod.py'>
On scripts#
Let us return to the diagnostics problem, and volmeans.py.
It is good that we can import volmeans and use it in Python, but we would
really like to be able to use this file as a script that we can run from
the terminal command line. We would like to be able to run the following
command, from the command line:
python3 volmeans.py
In our ideal world, this would print out the outliers for the scan
ds107_sub012_t1r2.nii.
We can do this very simply, by adding some stuff to the end of the module.
The addition is the print statement at the end of the module.
%%file volmeans.py
""" File to calculate volume means, detect outliers
Also, print out mean values.
"""
import numpy as np
import nipraxis
import nibabel as nib
import nipraxis
def vol_means(image_fname):
""" Calculate volume means from 4D image `image_fname`
"""
img = nib.load(image_fname)
data = img.get_fdata()
means = []
for i in range(data.shape[-1]):
vol = data[..., i]
means.append(np.mean(vol))
return np.array(means)
def detect_outliers_fixed(some_values, n_stds=2):
overall_mean = np.mean(some_values)
overall_std = np.std(some_values)
thresh = overall_std * n_stds
is_outlier = np.abs(some_values - overall_mean) > thresh
return np.where(is_outlier)[0]
# The new stuff:
data_fname = nipraxis.fetch_file('ds107_sub012_t1r2.nii')
means = vol_means(data_fname)
print(detect_outliers_fixed(means))
Overwriting volmeans.py
Now we can execute the .py file from the command line, to give the effect
we want.
Normally, to show this, you would open a terminal and type python3 volmeans.py. Here though, we will use the notebook ! prefix to ask the
notebook to execute code in the terminal. So, the notebook code below has
exactly the same effect as typing python3 volmeans.py at the terminal.
# Notice the ! at the beginning of the line below.
# This tells the notebook to execute the command in a terminal.
!python3 volmeans.py
[ 6 12 49 50 69 165]
There is another way to execute a Python file like this in a notebook,
which is to use the Jupyter %run command. This is particularly useful,
because we do not need to specify, for example, python3, because we are using
the Python session running inside the notebook.
%run volmeans.py
[ 6 12 49 50 69 165]
This is what we wanted, but…
The __name___ == "__main__" trick#
With the print at the end of our module, we have an annoying problem when we
import. As you remember, we have to reload the module to see the new
changes on import, but when we do…:
importlib.reload(volmeans)
[ 6 12 49 50 69 165]
<module 'volmeans' from '/home/runner/work/textbook/textbook/volmeans.py'>
Oh dear, we were doing the import to get access to the code, and it’s an annoying waste now, to calculate and print out the mean values when we do the import.
What we would really like to be able to do, is to calculate and print out the
values, only when we are executing the module as a script, and not when
we are importing the module to use the code.
Luckily Python gives us a way to tell whether the code is being run as part of a script or an import.
Before Python starts executing the code in the file, it sets a special variable, called __name__, that differs in the case of the executed script and the imported module.
Specifically:
When importing the code, the
__name__variable has a value that is a string containing the name of the module. In our case, when runningimport volmeans,__name__is the string'volmeans'.When executing the code, the
__name__variable has the value'__main__'.
See two underscores for more.
When you do want to use your .py file both as a script, and for
importing, you often see this pattern at the bottom of the .py file:
if __name__ == '__main__':
# We only get here if running the file as a script
print('I am running as a script')
See if name == ‘main’ in the Python documentation.
Now we can solve our problem using the volmeans.py for importing and as a script, like this:
%%file volmeans.py
""" File to calculate volume means, detect outliers
Also, print out mean values (only if running as a script).
"""
import numpy as np
import nibabel as nib
import nipraxis
def vol_means(image_fname):
""" Calculate volume means from 4D image `image_fname`
"""
img = nib.load(image_fname)
data = img.get_fdata()
means = []
for i in range(data.shape[-1]):
vol = data[..., i]
means.append(np.mean(vol))
return np.array(means)
def detect_outliers_fixed(some_values, n_stds=2):
overall_mean = np.mean(some_values)
overall_std = np.std(some_values)
thresh = overall_std * n_stds
is_outlier = np.abs(some_values - overall_mean) > thresh
return np.where(is_outlier)[0]
# The new stuff:
if __name__ == '__main__':
# The code is running as a script.
data_fname = nipraxis.fetch_file('ds107_sub012_t1r2.nii')
means = vol_means(data_fname)
print(detect_outliers_fixed(means))
Overwriting volmeans.py
# We do print out the values when running as a script:
%run volmeans.py
[ 6 12 49 50 69 165]
# We don't print the values when (re)importing:
importlib.reload(volmeans)
<module 'volmeans' from '/home/runner/work/textbook/textbook/volmeans.py'>
Command line arguments#
When you run a script, you can also pass command line arguments, e.g.:
python3 my_script.py a_string
Here my_script.py is the script, and a_string is the command line
argument.
You can do the same thing from within Jupyter / IPython:
run my_script.py a_string
In your script, you can get the command line arguments from the argv
list within the sys module. The first element of the sys.argv list is
always the name of the program – in our case this will be my_script.py.
The second and subsequent entries in sys.argv are the arguments entered at
the command line. For example, in our case:
sys.argv == ['my_script.py', 'a_string']
The entries in this list are always strings. For python3 my_script.py 1
you would get:
sys.argv == ['my_script.py', '1']
For example, to make volmeans.py more useful, we might want to pass a Nipraxis filename to the script, to get it to print out the means for any Nipraxis file, like this:
%%file volmeans.py
""" File to calculate volume means, detect outliers
When run as a script, print out values for given Nipraxis file.
"""
# New import of sys.
import sys
import numpy as np
import nibabel as nib
import nipraxis
def vol_means(image_fname):
""" Calculate volume means from 4D image `image_fname`
"""
img = nib.load(image_fname)
data = img.get_fdata()
means = []
for i in range(data.shape[-1]):
vol = data[..., i]
means.append(np.mean(vol))
return np.array(means)
def detect_outliers_fixed(some_values, n_stds=2):
overall_mean = np.mean(some_values)
overall_std = np.std(some_values)
thresh = overall_std * n_stds
is_outlier = np.abs(some_values - overall_mean) > thresh
return np.where(is_outlier)[0]
if __name__ == '__main__':
# New: using sys to get first argument.
nipraxis_fname = sys.argv[1]
data_fname = nipraxis.fetch_file(nipraxis_fname)
means = vol_means(data_fname)
print(detect_outliers_fixed(means))
Overwriting volmeans.py
Now we can run the script like this to print out the original set of means:
# Print out the values for ds107_sub012_t1r2.nii
%run volmeans.py ds107_sub012_t1r2.nii
[ 6 12 49 50 69 165]
We can also print out the mean values for any other Nipraxis file:
# Print out the values for ds114_sub009_t2r1.nii
%run volmeans.py ds114_sub009_t2r1.nii
[0]
See also#
Modules in the standard Python tutorial.




