Voxels and time#
See also: Reshaping, 4D to 2D.
# Import common modules
import numpy as np # the Python array package
import matplotlib.pyplot as plt # the Python plotting package
# Display array values to 6 digits of precision
np.set_printoptions(precision=4, suppress=True)
In this example, we calculate the mean across all voxels at each time point.
We’re working on ds114_sub009_t2r1.nii. This is a 4D FMRI image.
import nipraxis
bold_fname = nipraxis.fetch_file('ds114_sub009_t2r1.nii')
bold_fname
'/home/runner/.cache/nipraxis/0.5/ds114_sub009_t2r1.nii'
import nibabel as nib
img = nib.load(bold_fname)
img.shape
(64, 64, 30, 173)
We want to calculate the mean across all voxels. Remember that a voxel is a pixel with volume, and refers to a position in space. Therefore we have this number of voxels in each volume:
n_voxels = np.prod(img.shape[:-1])
n_voxels
122880
To calculate the mean across all voxels, for a single volume, we can do this:
data = img.get_fdata()
first_vol = data[..., 0]
np.mean(first_vol)
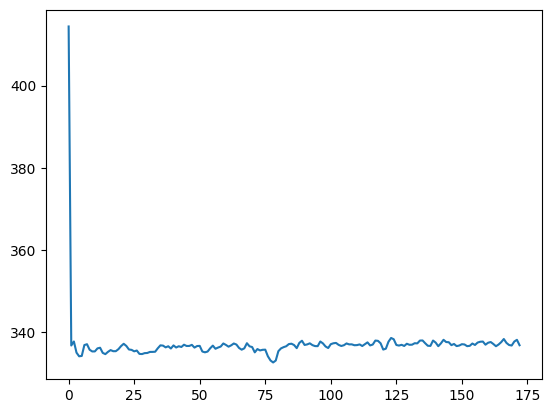
414.40107421875
To calculate the mean across voxels, we could loop across all volumes, and calculate the mean for each volume:
n_trs = img.shape[-1]
means = []
for vol_no in range(n_trs):
vol = data[..., vol_no]
means.append(np.mean(vol))
plt.plot(means)
[<matplotlib.lines.Line2D at 0x7fef28150a60>]
We could also flatten the three voxel axes out into one long voxel axis, using
reshape – see: Reshaping, 4D to 2D. Then we can use the axis parameter to
the np.mean function to calculate the mean across voxels, in one
shot. This is “vectorizing”, where we take an operation that needed a loop,
and use array operations to do the work instead:
voxels_by_time = np.reshape(data, (n_voxels, n_trs))
means_vectorized = np.mean(voxels_by_time, axis=0)
# The answer is the same, allowing for tiny variations.
assert np.allclose(means_vectorized, means)




